Meritshot Tutorials
- Home
- »
- R-Regular Expressions
R Tutorial
-
R-OverviewR-Overview
-
R Basic SyntaxR Basic Syntax
-
R Data TypesR Data Types
-
R-Data StructuresR-Data Structures
-
R-VariablesR-Variables
-
R-OperatorsR-Operators
-
R-StringsR-Strings
-
R-FunctionR-Function
-
R-ParametersR-Parameters
-
Arguments in R programmingArguments in R programming
-
R String MethodsR String Methods
-
R-Regular ExpressionsR-Regular Expressions
-
Loops in R-programmingLoops in R-programming
-
R-CSV FILESR-CSV FILES
-
Statistics in-RStatistics in-R
-
Probability in RProbability in R
-
Confidence Interval in RConfidence Interval in R
-
Hypothesis Testing in RHypothesis Testing in R
-
Correlation and Covariance in RCorrelation and Covariance in R
-
Probability Plots and Diagnostics in RProbability Plots and Diagnostics in R
-
Error Matrices in RError Matrices in R
-
Curves in R-Programming LanguageCurves in R-Programming Language
R-Regular Expressions
Regular expressions (regex) in R are a powerful tool for working with patterns in strings. They allow for complex search and manipulation of text based on specific patterns. In R, there are several functions designed to work with regex for tasks like searching, extracting, replacing, and splitting strings.
A regular expression, often denoted as regex or regexp, is a sequence of characters that defines a search pattern. It’s a powerful tool used in programming and text processing to search for and manipulate text based on specific patterns.
Using Regular Expressions in R
Here are some main functions that are used in Regular Expressions in R Programming Language.
Syntax:
- grepl()
- grepl(pattern, x) searches for matches of a pattern within a vector x and returns a logical vector indicating whether a match was found in each element.
- Checking if strings in a vector contain a specific
# grepl() returns the indices
grepL(pattern, x)
Example:
text <- “Hello, Meritshot!”
grepl(“Hello”, text)
Output:
TRUE
- sub() and gsub() – Replacing Patterns
- sub(): Replaces the first occurrence of a pattern in a
gsub(): Replaces all occurrences of a pattern in a string.
Syntax:
# sub() replaces the first occurrence of pattern
sub(pattern, replacement, x)
# gsub() replaces all occurrences of pattern gsub(pattern, replacement, x)
Example:
text <- “I love bananas and bananas are great.”
# Replace the first occurrence of “bananas”
new_text <- sub(“bananas”, “apples”, text)
print(new_text) # Output: “I love apples and bananas are great.”
# Replace all occurrences of “bananas”
new_text_all <- gsub(“bananas”, “apples”, text)
print(new_text_all) # Output: “I love apples and apples are great.”
3. regexpr() and gregexpr() – Finding Positions of Patterns
- regexpr(): Returns the starting position of the first match of a
- gregexpr(): Returns the starting positions of all matches of a
Syntax:
# regexpr returns the starting position of the first match regexpr(pattern, x)
# gregexpr returns the starting positions of all matches gregexpr(pattern, x)
Example:
text <- “The rain in India stays mainly in the plain.”
# Find the position of the first occurrence of “in”
position_first <- regexpr(“in”, text)
print(position_first) # Output: 6 (position of the first “in”)
# Find the positions of all occurrences of “in”
positions_all <- gregexpr(“in”, text)
print(positions_all) # Output: 6 15 36 45 (positions of all “in”)
4. strsplit() – Splitting Strings Based on Patterns
- strsplit(): Splits a string based on a regular expression (pattern) and returns a list of the
Syntax:
# strsplit() splits a string based on a pattern strsplit(x, split)
Example:
text <- “apple,banana,pear,grape”
# Split the string by commas
split_text <- strsplit(text, “,”)
print(split_text) # Output: List of “apple” “banana” “pear” “grape”
5. stringr Package – More User-Friendly String Manipulation Functions
The stringr package provides more intuitive and user-friendly functions for working with strings and regular expressions. It offers consistent function naming and argument handling.
To use the stringr package, you first need to install it:
install.packages(“stringr”)
Then, load the package:
library(stringr)
Key Functions in stringr for Regex:
- str_detect(): Equivalent to grepl(), returns TRUE or FALSE if a pattern is
- str_replace() and str_replace_all(): Equivalent to sub() and gsub().
- str_extract(): Extracts the first match of a pattern from a
- str_extract_all(): Extracts all matches of a pattern from a
- str_split(): Equivalent to strsplit() for splitting
Example:
# Using stringr functions
library(stringr)
text <- “apple, banana, cherry, grape”
# Detect if “apple” is in the string
is_apple <- str_detect(text, “apple”)
print(is_apple) # Output: TRUE
# Replace “banana” with “orange”
new_text <- str_replace(text, “banana”, “orange”)
print(new_text) # Output: “apple, orange, cherry, grape”
# Extract the first fruit (match before the first comma)
first_fruit <- str_extract(text, “[a-z]+”)
print(first_fruit) # Output: “apple”
Conclusion
Key Functions and Tools for Working with Regular Expressions in R: grepl() – Searching for Patterns
sub() and gsub() – Replacing Patterns
regexpr() and gregexpr() – Finding Positions of Patterns strsplit() – Splitting Strings Based on Patterns
stringr Package – A package offering more user-friendly regex functions
Common Regex Patterns in R
Regular expressions are defined using a series of symbols and patterns to match text in strings. Here are some of the most common regex patterns:
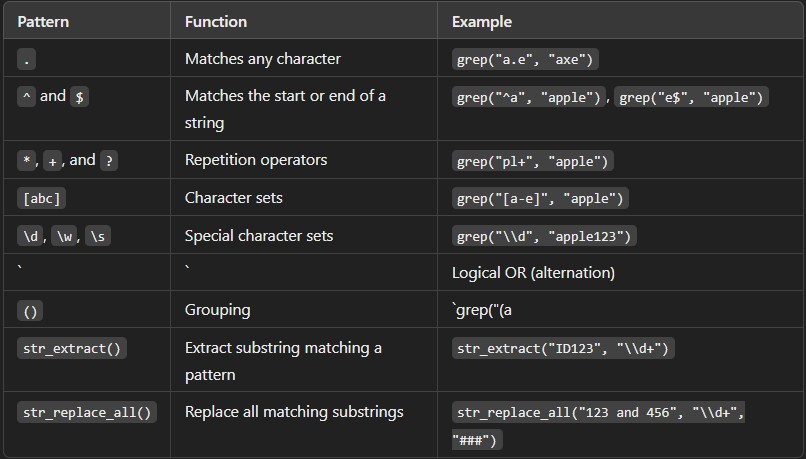
1. Using the Dot (.) to Match Any Character
The dot (.) is used to match any single character except for a newline (\n).
Example: Match Any Character
# Using grep() to find strings with any single character between “a” and “e”
text <- c(“axe”, “ape”, “apple”, “able”)
matches <- grep(“a.e”, text) # “.e” matches any character before “e”
print(text[matches]) # Output: “axe” “ape”
1. Using the Caret (^) and Dollar ($) for Anchors
- ^: Matches the start of the
- $: Matches the end of the
Example: Match the Start and End of a String
# Match strings that start with “a”
start_matches <- grep(“^a”, text)
print(text[start_matches]) # Output: “axe” “ape” “apple” “able”
# Match strings that end with “e”
end_matches <- grep(“e$”, text)
print(text[end_matches])
# Output: “axe” “ape” “apple”
3. Using *, +, and ? for Repetition
- *: Matches 0 or more occurrences of the preceding
- +: Matches 1 or more occurrences of the preceding character.
- ?: Matches 0 or 1 occurrence of the preceding
Example: Repetition Patterns
# Match strings with “p” followed by 0 or more “1”
zero_or_more <- grep(“pl*”, text)
print(text[zero_or_more])
# Output: “apple”
# Match strings with “p” followed by 1 or more “l”
one_or_more <- grep(“pl+”, text)
print(text[one_or_more]) # Output: “apple” # Match strings with “a” followed by 0 or 1 “p” zero_or_one <- grep(“ap?”, text) print(text[zero_or_one])
# Output: “ape” “apple”
4. Using Brackets ([]) for Character Sets
- [abc]: Matches any one of the characters a, b, or
- [a-z]: Matches any lowercase letter in the range from a to
Example: Character Sets
# Match strings containing any letter from “a” to “e”
char_set <- grep(“[a-e]”, text)
print(text[char_set])
# Output: “axe” “ape” “apple” “able”
1. Using Backslash (\) for Special Characters
- \d: Matches any digit (0-9).
- \w: Matches any word character (letter, digit, or underscore).
- \s: Matches any whitespace character (space, tab, newline).
Example: Special Characters
# Matching strings containing any digit
text_with_digits <- c(“car123”, “plane”, “boat9”, “train”)
digit_match <- grep(“\\d”, text_with_digits)
# Escape \d as \\d in R
print(text_with_digits[digit_match]) # Output: “car123” “boat9”
# Match strings with a word boundary for “boat”
word_boundary <- grep(“\\bboat\\b”, text_with_digits) print(text_with_digits[word_boundary]) # Output: “boat9”
1. Using | for Alternation (Logical OR)
The vertical bar (|) is used to create an OR condition between patterns.
Example: Match Multiple Patterns
# Match strings that contain “apple” or “ape”
or_match <- grep(“apple|ape”, text)
print(text[or_match])
# Output: “ape” “apple”
7. Using Parentheses (()) for Grouping
Parentheses are used to group parts of a pattern together, often for repetition or alternation.
Example: Grouping Patterns
# Match “a” followed by either “p” or “x”
grouping_match <- grep(“a(p|x)”, text)
print(text[grouping_match])
# Output: “axe” “ape” “apple”
8. Using str_extract() to Extract a Substring
With the stringr package, you can extract substrings that match a regex pattern using str_extract().
Example: Extract Substrings
library(stringr)
# Extract the first occurrence of a digit in each string
strs <- c(“ID123”, “code456”, “item789”)
extracted_digits <- str_extract(strs, “\\d+”)
print(extracted_digits)
# Output: “123” “456” “789”
9. Using str_replace() and str_replace_all() for Replacement
To replace parts of a string based on a regex pattern, use str_replace() (for the first match) or str_replace_all() (for all matches).
Example: Replace Patterns
# Replace the first occurrence of a number with “###”
replaced_str <- str_replace(“car123”, “\\d+”, “###”)
print(replaced_str) # Output: “car###”
# Replace all occurrences of numbers with “###”
replaced_all_str <- str_replace_all(“car123 and boat456”, “\\d+”, “###”) print(replaced_all_str) # Output: “car### and boat###”
Summary of Using Patterns with Functions:
Regular expressions are a highly versatile tool in R, enabling precise and complex text manipulations. You can use them in combination with functions like grep(), sub(), and stringr to perform advanced searches, replacements, and extractions in text data.




